Wanted to share my final paper for bioc218, a computational biology class i took this winter 2013. We were tasked with choosing a particular technique, algorithm, or any material discussed in the class and exploring it in more detail. Neural networks seemed particularly interesting—and not just because i'm a neuroscientist. I wanted to use machine learning algorithms to improve predictions for the project i am working on in Judith Frydman's lab.
For those who want to dive right in, the paper:
bioc218 final paper
Wanted to share my final paper for bioc218, a computational biology class i took this winter 2013. We were tasked with choosing a particular technique, algorithm, or any material discussed in the class and exploring it in more detail. Neural networks seemed particularly interesting—and not just because i'm a neuroscientist. I wanted to use machine learning algorithms to improve predictions for the project i am working on in Judith Frydman's lab.
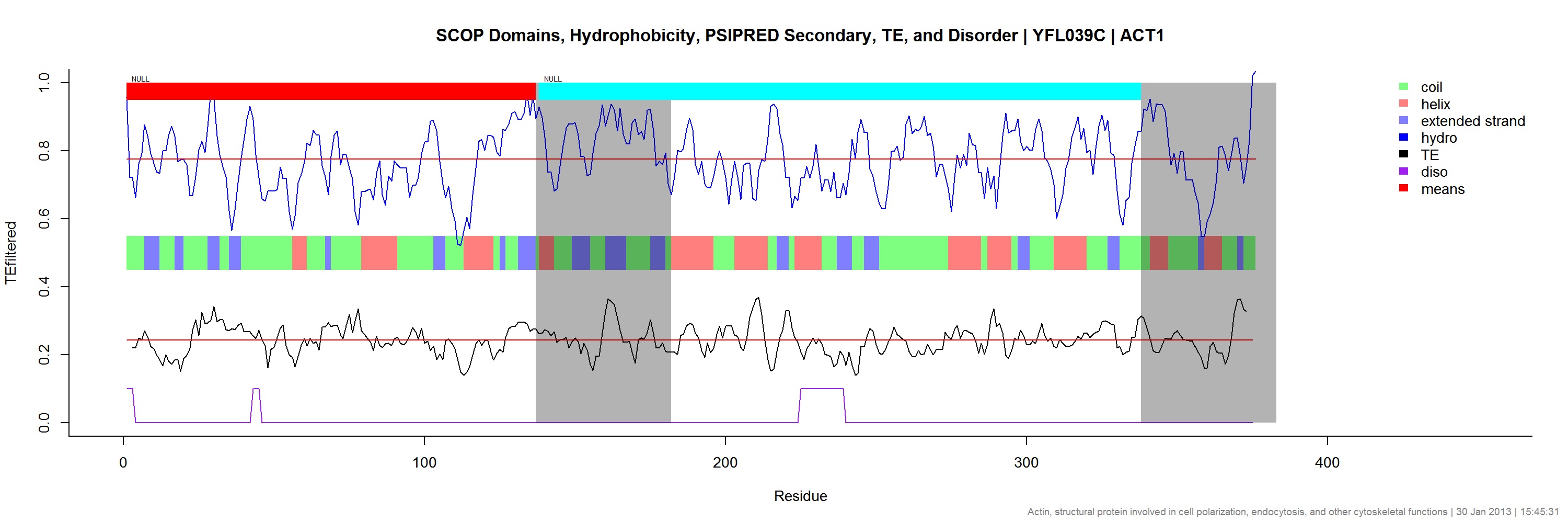
We are using codon optimality to predict sites of ribosome pausing, which could help facilitate proper protein domain folding. This would be validated with ribosomal profiling or other emerging techniques. The higher-level idea is that synonymous substitutions in a protein coding region could affect an organism's fitness, as recently shown with Synechococcus elongate's kaiBC and Neurospora's FRQ. This mechanism has also been seen to play a role in MDR1 induced cancer and it is possible that other protein folding diseases, especially neurodegenerative ones, might be linked by this common pathway. Elucidating it could lead to novel therapies or reveal basic biological mechanisms.
The final report encompasses both a description of the history and implementation of neural networks. In addition, i went beyond the requirement by proposing and demonstrating initial analysis for applying neural networks to discovering novel sites of domain specific ribosomal pausing in the genome. While the results indicate much works needs to be done, the conceptual framework is there to continue. Further, i included a short section proposing an extension to current neural network methods that is subsequently shown to be valid through an article i discuss briefly. The paper was super fun to write (in LaTeX of course!) and i hope to finish the project at some point this summer.
 stanford
stanford linkden
linkden github
github goodreads
goodreads medium
medium twitter
twitter